Analyzing protein-protein spatial-temporal dependencies from TIRFM image sequences using temporal random sets
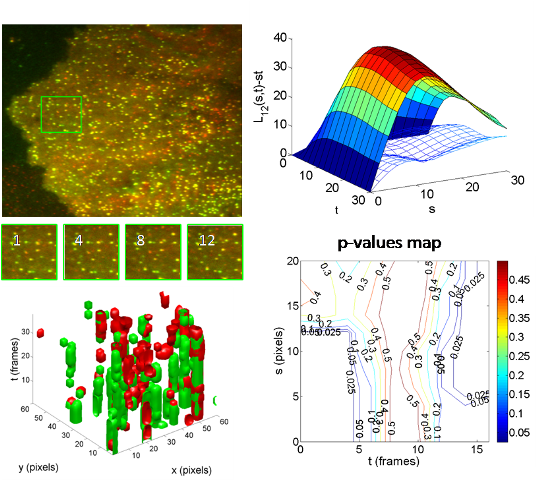
Recent advances in microscopy allow to image dynamic
processes with a very high spatial and temporal resolution. Using TIRFM
imaging of GFP-tagged clathrin endocytic proteins, areas of
fluorescence are observed as overlapping spots of different sizes
and durations forming random clumps (see Figure). Standard procedures to measure protein-protein
colocalization of dual labeled samples threshold the original
gray-level images to segment areas covered by different proteins.
This binary logic is not appropriate as it leaves a free tuning
parameter which can influence the conclusions. Moreover, these
procedures rely on simple statistical analysis based on
correlation coefficients or visual inspection.
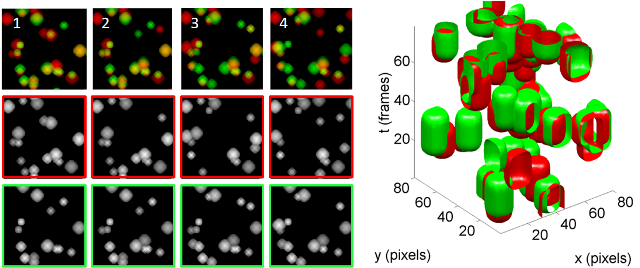
We propose a probabilistic approach. We model the image sequences of two proteins as a realization of a bivariate fuzzy temporal random set. We have developped a
non-parametric approach to quantify spatial and spatio-temporal
interrelations by using the pair-correlation function, the
cross-covariance function and the Ripley K-function. Based on these summary statistics, we use a randomization procedure
to test independence between types by applying random toroidal
shifts and a Montecarlo test. Our results show
these statistics accurately capture spatio-temporal dependencies.
Estimation of the interaction range is also obtained. The method has
been successfully applied to analyze relationships between endocytic
proteins from fluorescent-labeled proteins image sequences involved in
endocytosis (Clathrin, Hip1R, Epsin and Caveolin). This procedure allows biologists to automatically quantify dependencies between molecules in a more formal and robust way.
Software
Temporal Random Sets. A Matlab toolbox for simulation and estimation
Publications
- Díaz ME, Ayala G, León T, Zoncu R, Toomre D. Analyzing protein-protein spatial-temporal dependencies from image sequences using fuzzy temporal random sets. Journal of Computational Biology. 15(9):1221-36, 2008.
- Díaz E, Sebastian R, Ayala G, Díaz ME, Zoncu R, Toomre D, Gasman S. Measuring spatial temporal dependencies in bivariate random sets with applications to cell biology. IEEE Transactions on Pattern Analysis and Machine Intelligence. 30(9):1659-71, 2008
- Ayala G, Sebastian R, Díaz ME, Díaz E, Zoncu R, Toomre D. Analysis of spatially and temporally overlapping events with applications to image sequences. IEEE Transactions on Pattern Analysis and Machine Intelligence. 28(10):1707-12, 2006
- Sebastian R, Díaz E, Ayala G, Díaz ME, Zoncu R, Toomre D. Studying endocytosis in space and time by means of temporal Boolean models. Pattern Recognition. 39(11):2775-85, 2006

|
|