BFU2015-65037-P
Title: Virus evolution in hosts of varying susceptibility: fitness and virulence consequences of the evolution of host-virus protein-protein interaction networks
Research Group: Evolutionary Systems Virology
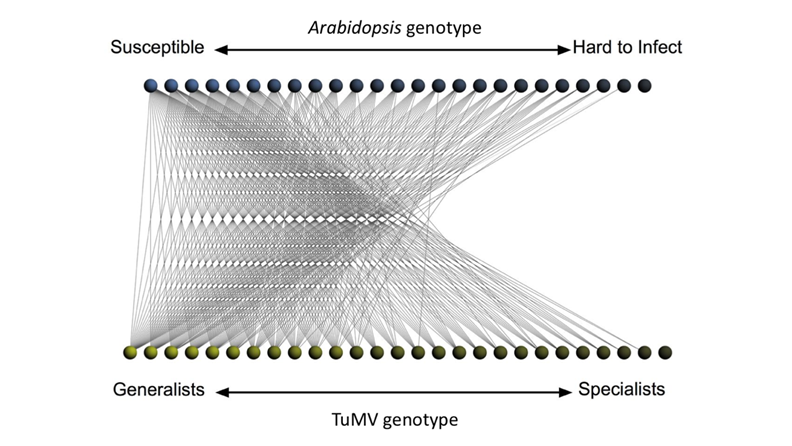
It is generally assumed that genetic variability in host species for susceptibility to infection will necessarily condition the evolution of pathogens populations, either by driving them to the diversification of the pathogen into strains that track the different host defence alleles (e.g., antigenic diversity), or by canalization of the pathogen to infect only the most susceptible genotypes. Associated to these processes of diversification or specialization, virulence may or may not increase concomitantly. In any case, pathogen's fitness must be optimized.
However, these expectations have been poorly tested experimentally for RNA viruses, specially the evolutionary implications. Here, we propose a large scale evolution experiment in which independent lineages of turnip mosaic potyvirus (TuMV) will be evolved in a number of different genotypes of Arabidopsis thaliana ecotype Col-0 that differ in their degree of susceptibility to infection with this virus. The collection comprises genotypes with mutations in several defence pathways that either lead to increased or decreased responses to infection, as well as in other plant genes that are known to be essential for viral infection. After experimental evolution, we will characterize the virus population diversity throughout the evolution experiment as well as the fitness and virulence of the endpoint populations. Next, we will test whether adaptation to a given host genotype is specific or, by contrast, viruses evolved as generalists. Finally, we will sought for the molecular basis of the adaptation process in terms of changes in the interactions between the host proteome and each of the evolved viral proteins. To do so, we will use a high throughput screening of protein-protein interactions using a commercial yeast two hybrids system. We will construct host-virus protein-protein interaction networks for each evolved viral protein and compare in silico the effect that gains or losses of interactions by these proteins may have in the network. We will correlate such changes with the observed phenotypic changes (virulence and viral fitness).
Ref. BFU2015-65037-P
Santiago F. Elena
- Silvia Ambrós
- Rubén González
Agencia Estatal de Investigación, Ministerio de Ciencia, Universidades e Investigación - FEDER
- MCiencia - Innovation