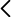EPICOVIR
Title: The role of host’ epicode on the evolution of viral populations
Research group: Evolutionary Systems Virology and Theory, Bioinformatics and Computation
The generation of mitotically stable alterations in gene expression due to epigenetic marks is a fast and relatively long-lasting manner for stablishing a genomic memory of past stress events. Environment-triggered deregulation of genetic factors associated with epigenetic machineries can also lead to phenotypic plasticity and stress mitigation. Therefore, host’s epigenetic machinery can pose an important but largely unmeasured selective pressure on pathogens. Plant viruses offer a convenient model for studying this kind of interactions. Firstly, a large-scale evolution experiment for checking how virus populations evolve and interact in plants with compromised or enhanced epigenetic pathways is proposed. Arabidopsis thaliana plants with mutations in key genes associated with active or repressive chromatin marks, including DNA methylation and histone modification, will be challenged against independent lineages of turnip mosaic potyvirus (TuMV). Plant genotypes having different degree of susceptibility to TuMV will be repeatedly inoculated with the virus and measures of virulence, fitness and diversity of viral populations will be assessed during specific time points and the contribution of selection and drift to evolution evaluated. Secondly, the antiviral effect of epigenetic modifications in plants infected with TuMV will be tested in the context of (i) superinfection of the same plants and (ii) transgenerational protection against future infections. For these experiments, viruses of decreasing degree of genetic relatedness with TuMV will be used to prime plants that will be then inoculated with TuMV. An Integrative Systems Biology approach will be used for networking the viral effects on the expression of host’s transcripts, small RNAs and genome-wide methylation profiles. The molecular basis of the adaptive process will be tested by constructing host-virus protein-protein interaction networks for each evolved viral protein, followed by in silico analysis of the observed interactors. Results cannot only potentially identify epigenetic pathways playing important roles in the responses against viruses, but also shed light on the impact of these chromatin regulators on the evolution of viral populations. Furthermore, by using genotypes with varying susceptibility to virus infection, results can possibly contribute for understanding the rules dictating the fate of epidemics associated with emerging viruses, an issue with social and economic relevance.
Ref. PID2019-103998GB-I00
Santiago F. Elena
- Vicente Arnau
- Régis L. Corrêa
- Denis Kutnjak
Agencia Estatal de Investigación - FEDER