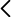Type: Equipment
The Array platform, designed by Affymetrix, provides a holistic study of organisms from the perspective of systems biology, a better understanding of biological properties and interactions as a whole. This technology provides the flexibility to study a specific subset of genes of interest or to be able to focus on the study of the genome at a global level in order to understand and define in depth the complex mechanisms and networks that underlie biological processes and diseases.
The Service has the most advanced microarray system designed by the leading company Affymetrix. This system, called Genechip®, provides an effective, reliable and reproducible methodology to obtain the gene information.
The Array platform, with modern technology and powerful instrumentation, enables the simultaneous analysis of thousands of genes quantitatively and reproducible, offering an optimal balance of sensitivity and specificity for an accurate evaluation of expression, for any number of genes in any model of organism.
This integrated platform enables a wide range of studies to be carried out experiments to get a complete view of biological systems. Offers a large selection of matrices for studying organisms from Arabidopsis up to Zebrafish, in addition to custom options with CustomExpress, which allows to design your own array with the parameters and sequences of interest.
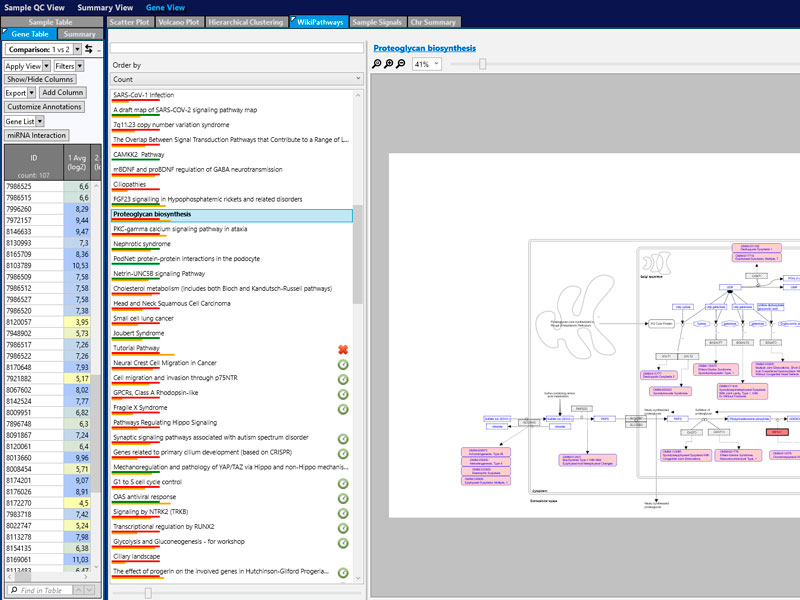
Arrays have been applied to the study of almost any type of biological problem. The number of annual publications is very high and continues to grow. Its purposes include discovering new regulatory pathways, mechanisms action confirmation, drug target validation, classification of diseases, response to toxicological analysis and development of diagnosis.
Among the most frequent, there are:
- Study of genes that are distinctly expressed under distinct conditions (healthy/sick, mutant/wild, treated/untreated).
- Molecular classification in complex diseases. Identification of characteristic genes of a pathology (signature).
- Prediction of response to a treatment.
- Detection of mutations and single gene polymorphisms (SNP).
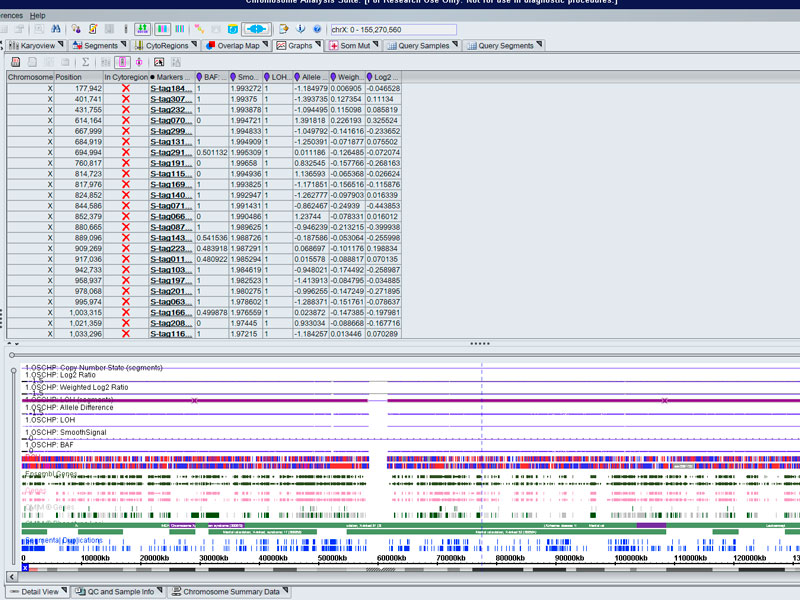
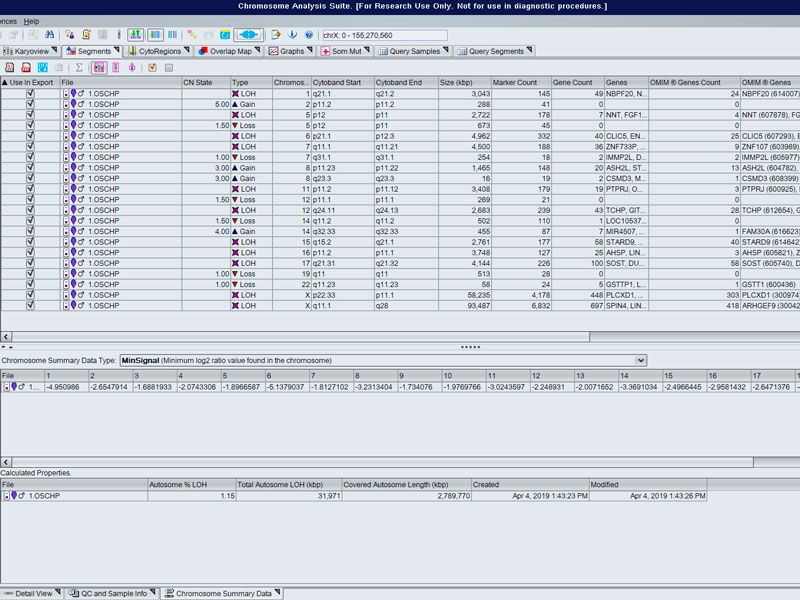
- Detection of chromosomal aberrations: CNV / SNPs, LOH, mosaicism, UDP...
- Array for the study of traditional gene expression using 3’ IVT Expression Analysis. For conventional analysis of patterns of expression.
- Array for the study of the complete transcriptome Gene Arrays/Clariom S for studies of differential expression in greater depth by contemplating transcripts without PolyA ends: lncRNA, lincRNA, etc.
- Array for studying splicing variants using the new Clariom D/Transcriptome Array and alternatively Exon Array. For studies of gene expression levels and exon levels.
- Array for the study of small-RNA, for studies of mechanisms mediated by miRNAs, pre-miRNA, snoRNA, scaRNA.
- Array for transcriptomic mapping Whole Genome Arrays covering whole genome: study of gene expression, alternative splicing, miRNA….
- Array for studying DNA/protein interactions (ChIP-On-Chip) with different types of arrays of total coverage (Tiling Array). Promoters.
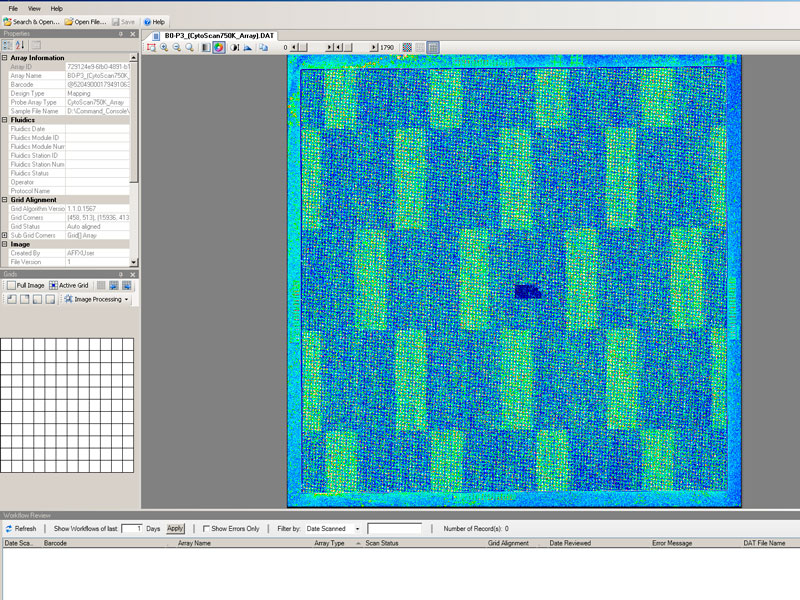
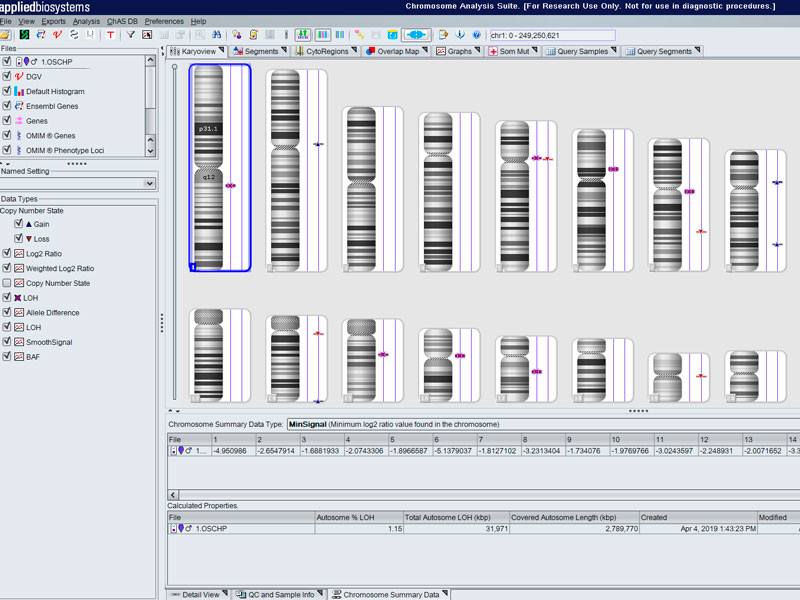
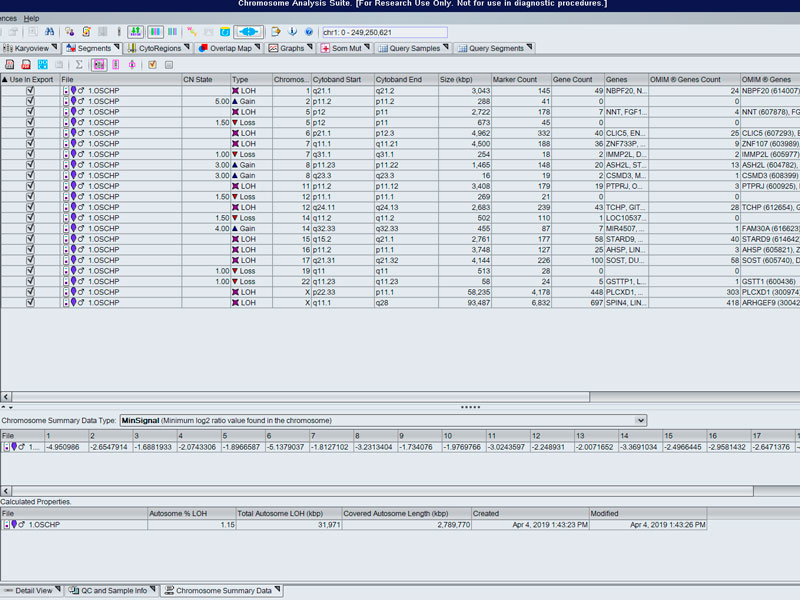
- Array for the study of molecular cytogenetics with high sensitivity, specificity and resolution for the identification of chromosomal aberrations in quality control of cell lines (stem cell incl.), prenatal studies tumour cells... in the detection of abnormalities, aberrations submicroscopic, loss/absence of heterozygosity, gains and losses, uniparental and mosaic dysomias.
The interested user can contact the staff of the service to request information and/or budget. The budget should always be signed by both parties, both by the person in charge of the project and by the Coordinator of the UCIM.
Once the budget is accepted, the user will provide the samples* for the selected study.
*Recommendations for samples:
- 3 replicates per experimental condition.
- Recommended, for any extraction kit, to elute the sample (RNA/DNA) in nuclease-free water.
- The required sample concentration will depend on the protocol. There is no inconvenience in sending more or more sample quantity concentrated, since the Service will be able to normalize the samples before the start of the protocol.
- The user will provide a high quality and purity sample. Before the beginning of the study, the corresponding quality control methods are going to be applied to the selected sample. The user will be informed of those that do not meet of the minimum of quality to look for possible alternatives and/or solutions. The volume of sample left over after the study may be returned to the user if requested.
- El volumen de muestra sobrante tras la realización del estudio podrá ser devuelto al usuario si así lo solicita.
- Regarding the shipment of samples, they must be well labelled and identified, and accompanied by an explanatory sheet describing the data of the samples.
- RNA/DNA samples will be shipped frozen in dry ice at 1.5 mL tubes in nuclease-free water.
The results will be delivered by email, through a download link, within the indicated period. Or through the option chosen by both parties.
Upon receipt of the results, the user must sign a document provided by the Service to confirm the reception of the mentioned results.
- Portero Trigo, Jesica Maria
- PIT-Investigacio Escala Tecnica Superior