THE POLAR SOLAR PANELS MICROBIOME
EL MICROBIOMA DE LOS PANELES SOLARES
Abstract: Solar panels located on high (Arctic and Antarctic) latitudes combine the harshness of the climate with that of the solar exposure. We report here that polar solar panels are inhabited by similar microbial communities in taxonomic terms, dominated by Hymenobacter spp., Sphingomonas spp., and Ascomycota. Our results suggest that solar panels, even on high latitudes, can shape a microbial ecosystem adapted to irradiation and desiccation.
Article published! ¡Artículo publicado! Polar solar panels: Arctic and Antarctic microbiomes display similar taxonomic profiles, Dec 2017, Environmental Microbiology, DOI 10.1111/1758-2229.12608.
Resumen: Los paneles solares localizados en latitudes elevadas (Ártico y Antártico) combinan la dureza de la exposición solar con la del clima. Hemos descubierto que los paneles solares están habitados por comunidades microbianas similares en términos taxonómicos, dominadas por Hymenobacter spp., Sphingomonas spp., y Ascomycota. Nuestros resultados sugieren que los paneles solares, incluso en latitudes elevadas, pueden albergar un ecosistema microbiano adaptado a la irradiación y la desecación.
Methods overview
Sampling
Sampling of five solar panels on the rooftop of the technology building of the University of Tromsø (69.6815 N 18.9759 E) was performed in May 2017 (air temperature 3-6 °C). Two vertical panels attached to the wall (98 x 69 cm) and three inclined panels (50-60°) next to each other (170 x 98 cm) were sampled. All of the panels had a southwards orientation and some displayed visible bird depositions that were removed before sampling.
Samples were also taken from solar panels on two Antarctic islands, Livingston (62°39′S, 60°23′W) and Deception (62°58′S, 60°40′W), where presence of solar panels is linked to the activity of Scientific Spanish Bases. Livingston Island (850 km2) is the second largest island of the archipelago and is mostly occupied by glaciers. Deception Island (72 km2), with 57 % of the island covered by permanent glaciers, is an active volcano with recent eruptions and continuous fumarolic activity. On January 2017, three solar panels (isolated and dispersed around the Spanish Bases) were sampled at each of the islands. Panels were sized from 45 x 40 cm to 45 x 80 cm, were totally vertical or very slightly inclined, displayed a northwards orientation, and no bird depositions were visually detected.
In all cases, sampling of the solar panels was performed by strongly and repeatedly scratching the surface with a sterile window cleaners soaked with phosphate saline buffer (PBS). The resulting liquid was collected using sterile pipettes and was transferred to sterile falcon tubes that were kept on ice until frozen at -20 °C. The samples were then shipped to Valencia, Spain where they were further analyzed.
Shotgun metagenomics: DNA extraction, sequencing, computation and analysis
A 2 mL aliquot of the solar panel samples was separated for culturing assays, and the remaining volume was centrifuged at 4500 rpm for 30 minutes and concentrated to a volume of 500 µL per sample. DNA extraction was performed using the Power Soil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA, USA), quantified with Nanodrop-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and sequenced at Lifesequencing SL (Valencia, Spain) on a NextSeq500 (Illumina) sequencer.
Pre-analysis
Shotgun metagenomic sequencing data was quality checked using FastQC v0.11.5 (Babraham Bioinformatics, 2016) and MultiQC v1.0 (Ewels et al. 2016).
Classification
The metagenomic sequences were analyzed with the Centrifuge software package (Kim et al., 2016) version 1.0.3-beta (Dec 2016), run in parallel within a shared-memory fat node, using 60 threads and peaking half a tebibyte of DRAM. The database used for Centrifuge was generated in-house from the NCBI whole genomes nt database (nucleotide sequence database, with entries from all traditional divisions of GenBank, EMBL, and DDBJ) and index databases, downloaded in Aug 2017. Once generated, it was 15% larger than the last public available Centrifuge nt database of Nov 2016.
Post-analysis
The results generated with Centrifuge were visualized using Pavian (Breitwieser & Salzberg, 2016). All this pipeline was also used to reanalyze the SMS data from Dorado-Morales et al, 2016. Further deeper analysis and comparative metagenomics for all the samples analyzed was performed through the Aug 2017’s release of the Recentrifuge software (Martí, 2017), which enabled not only scoring each leaf in the classification tree, but also extracting common and exclusive taxa at different taxonomic levels. A comprehensive set of interactive plots is available in the following section (Comparative Metagenomics Analysis).
Isolation of microorganisms and 16 rRNA identification
An aliquot (100 µL) of each solar panel sample was cultured on LB (composition in g/L: peptone 10.0, NaCl 10.0, yeast extract 5.0, agar 15.0) and R2A (composition in g/L: peptone 0.5, yeast extract 0.5, dextrose 0.5, soluble starch 0.5, K2HPO4 0.3, MgSO4 0.05, sodium pyruvate 0.3) medium and the plates were incubated for two weeks at 4°C. A total of 44 different colonies were selected, re-streaked several times for purification and conserved in 20% glycerol at -80°C for further uses.
For 16S rRNA identification, a 500-bp fragment of the hypervariable region V1-V3 of the isolates was amplified by colony PCR, using universal primers 28F (5′ -GAG TTT GAT CNT GGC TCA G-3′) and 519R (5′ -GTN TTA CNG CGG CKG CTG-3′). Amplicons were checked in 1.4% agarose gel and precipitated overnight in isopropanol 1:1 (vol:vol) and potassium acetate 3M pH 5 1:10 (vol:vol). Precipitated DNA was washed with 70% ethanol, resuspended in Milli-Q water (Merck Millipore Ltd, Tullagreen, Cork, Ireland) and quantified using a Nanodrop-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Amplicons were tagged using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA) and subjected to Sanger sequencing by the sequencing service of the University of Valencia (Spain). The resulting sequences were manually edited using Pregap4 (Staden Package, 2002) to eliminate low-quality base calls.
Stress-resistance assays
For temperature-resistance assays, 5 µL drops of a suspension of each isolate were placed on LB or R2A medium (supplemented with 20% glycerol in the case of incubation at -15 °C) and the plates were incubated at different temperatures (-15, 4, 15, 30, 37 and 50 °C) for several days.
UV-radiation resistance was tested by plating 5 µL drops of a suspension of each isolate (OD600 0.03) on LB or R2A medium, and by irradiating these plates from 15 cm with UV pulses with a Vilber VL-4.C lamp (254 nm, 340 μW/cm2; Labolan, S.L., Spain) for 30 seconds, 2 minutes or 8 minutes. The wavelength was chosen to be in the proximity to both the peak of the absorption spectrum of DNA and the peak of the absorption spectrum in ozone, which are very close one to the another. After irradiation plates were incubated at 15 °C and growth was assessed after 5 days.
Finally, desiccation-resistance was studied by adding 5 µL drops of a suspension of each isolate (OD600 0.3) on sterile filter paper, which was then dried in sterile conditions at 25 °C for 4 hours, followed by a transference of the microorganisms to fresh medium and incubation at 15 °C for five days, after which growth was assessed.
References
- Breitwieser FP, Salzberg SL (2016). Pavian: Interactive analysis of metagenomics data for microbiomics and pathogen identification. bioRxiv: 084715.
- Ewels P, Magnusson M, Lundin S, Käller M (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32(19): 3047-3048.
- Kim D, Song L, Breitwieser FP, Salzberg SL (2016). Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26:1721-1729.
- Martí JM (2017). Recentrifuge: robust comparative analysis and contamination removal for metagenomic data. bioRxiv: 190934.
Comparative Metagenomics Analysis
Results
Recentrifuge plots
Results after classification with Centrifuge and analysis with Recentrifuge:
| MHL | Artic Recentrifuge plots | Antarctic Recentrifuge plots | Polar Recentrifuge plots | All samples Recentrifuge plots | MHL |
|---|---|---|---|---|---|
| 20 | Artic MHL=20 | Antarctic MHL=20 | Poles MHL=20 | All MHL=20 | 20 |
| 22 | Artic MHL=22 | Antarctic MHL=22 | Poles MHL=22 | All MHL=22 | 22 |
| 25 | Artic MHL=25 | Antarctic MHL=25 | Poles MHL=25 | All MHL=25 | 25 |
| 30 | Artic MHL=30 | Antarctic MHL=30 | Poles MHL=30 | All MHL=30 | 30 |
| 35 | Artic MHL=35 | Antarctic MHL=35 | Poles MHL=35 | All MHL=35 | 35 |
| 40 | Artic MHL=40 | Antarctic MHL=40 | Poles MHL=40 | All MHL=40 | 40 |
| MHL = Minimum Hit Length | |||||
Genera relative frequencies
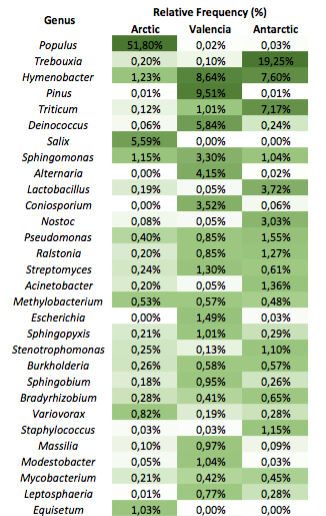

This heatmap is showing the 30 most abundant genera versus geographical location ordered by overall relative frequency (the average for all the solar panels sampled per location is given). The color scale is quasi-logarithmic to improve visualization of taxa with similar order of magnitude among locations. Unlike Figure 2D in the paper, the taxa under the clade Streptophyta have NOT been removed.
Further details
Excluded taxa from the recentrifuge plots
List of taxa (and below) to be excluded:
TaxId Scientific Name
7711 Chordata
12908 unclassified sequences
1747 Cutibacterium acnes
(1912216 Cutibacterium)
860235 Kibdelosporangium phytohabitans
28384 other sequences
Contact
For more information, please contact:

I2SysBio Synthetic Biology Laboratory
Institute for Integrative Systems Biology - 4th floor

