Spanish version
| |
|
Spanish version |
|||||
|
|
|||||
|
|
|||||
|
Exploration of the actin cytoskeleton of Saccharomyces cerevisiae
|
1. Input data |
|
For this example, a set of 34 Saccharomyces cerevisiae proteins charaterized by Drees et al. (2001) will be used. It comprises 26 proteins participating in actin patch assembly and patch-mediated endocytosis, together with eight proteins involved in other related proceses. Results shown in that study were updated by analyzing the DIP database to generate the following graph of protein-protein interactions (drawn using PIVOT). |

| After processing these
primary interaction data with UVCLUSTER, twelve clusters could be
determined (for details on the choosing of the partition, see
Arnau et al.
(2004)).
|
 |
 |
|
Biological validation of the clusters through SGD Gene Ontology Term Finder |
| Cluster |
Proteins |
GO Term with the lowest probability of cluster occurring by chance |
p[cluster] |
| 1 |
SLA1 RSV167 YSC84 SLA2 ABP1 YOR284w YGR268c |
Actin cytoskeleton organization and biogenesis |
3.01·10-11 |
| 2 |
YPL246c LAS17 YHR133c |
No significant ontology term found |
|
| 3 |
ACF2 |
The program requires at least two proteins in a cluster |
|
| 4 |
YJR083c |
The program requires at least two proteins in a cluster |
|
| 5 |
RVS161 YBR108w |
No significant ontology term found |
|
| 6 |
CDC42 CLA4 GIC2 |
Rho protein signal transduction |
1.51·10-8 |
| 7 |
CAP1 CAP2 YPR171w |
Actin cytoskeleton organization and biogenesis |
2.08·10-6 |
| 8 |
SWE1 HSL7 APP1 |
G2/M transition of mytotic cell cycle |
6.15·10-5 |
| 9 |
CRN1 SVL3 |
Cell growth and/or maintenance |
0.09242 |
| 10 |
BNI1 PFY1 BNR1 |
Response to osmotic stress |
5.06·10-7 |
| 11 |
TRM5 ACT1 SRV2 AIP1 COF1 |
Actin filament depolymerization |
7.55·10-7 |
| 12 |
YNL086w |
The program requires at least two proteins in a cluster |
|
|
|
|
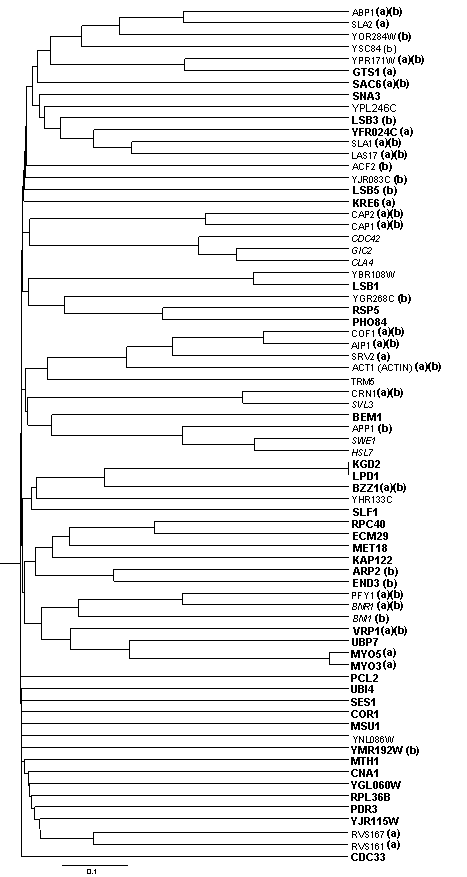
Incluiding all the avalaible data from S.cerevisiae using UVCLUSTER allows to investigate wether there are other proteins that may be significantly involved in actin patch assembly and function. In order to do so, the average primary distance of each of the proteins in the DIP dataset (4721 by the time of the study) against the 26 proteins involved in actin patch assembly and function according to Drees et al. (2001) (compraising clusters 1-5, 7 and 11-12) is calculated. That can be acomplished providing UVCLUSTER with the full list of proteins, leaving the ones of interest at the begining. The output file S1 includes the primary distances table that can be exported to an spreadsheet. Only the first colums, corresponding to the proteins of interest, are significant. 19 of the 26 proteins were among the 40 proteins with lowest average distances in the whole dataset (ranging from 1.31 to 2.23). The worst connected protein (TMR5) was in position 199 (2.65). The UPGMA tree was obtained using the whole DIP interaction data, with AC=100 and N=10000 and selecting the original list of proteins plus 38 other proteins potentially involved in actin patch assembly and function (according to the detailed procedure; distance lower than 2.27). The new proteins are highlated in bold. (a): Proteins that are localized to the actin cytoskeleton according to Huh et al. (2003). (b): Proteins assigned to the GO process "actin cytoskeleton and biogenesis" according to the SGD database. As can be seen, the new proteins distribute themselves among the previously determined clusters with only five old ones appearing in very different possition.
|
 |