|
|||||
Home Publications
|
Virus evolution lab
Research tools
Experimental
evolution
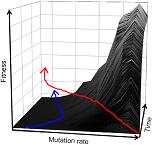
We use evolution in the lab to test evolutionary hypotheses and to direct traits of interest towards predefined goals for applied purposes (directed evolution). To achieve these goals, we have used bacteriophages, vesicular stomatitis virus (VSV), HIV-1, hepatitis C virus, enteroviruses, viroids, and adenoviruses, among other viruses. We have also implemented the use of VSV recombinant viruses expressing the receptor binding proteins of other enveloped viruses. Molecular and cell biology The techniques we are currently using include massive parallel sequencing (including standard and high-fidelity sequencing and RNA-seq), genetic engineering and gene synthesis, site-directed mutagenesis, RT-qPCR, flow cytometry, single-cell analysis, automated quantitative microscopy, and electron microscopy, among others. Comparative biology The comparison of species is a classic approach in biology. We combine this approach with experimentation. One of our main research goals is to compare the evolutionary properties of different viral species, including their ability to generate genetic variation, adapt to novel environments, tolerate deleterious mutations, or evolve new functional capabilities in the laboratory. Computational biology and modeling We use computational tools for phylogenetic analysis, sequence analysis, modeling, etc. We also use machine learning and deep learning models to make predictions about viral tropism. Universitat de València, 2025
|