The Membrane Biophysics Group
Latest News
04 January 2024
Open Call
Interested in conducting a PhD in the topic of optical and chemical control of membranes, peptides and proteins?
About us
We are a multidisciplinary group (see Members) working at the Institute of Molecular Science - ICMol (Scientific Park of the University of Valencia).
Please, visit the Contact page if you want to get in touch with us.
What we do
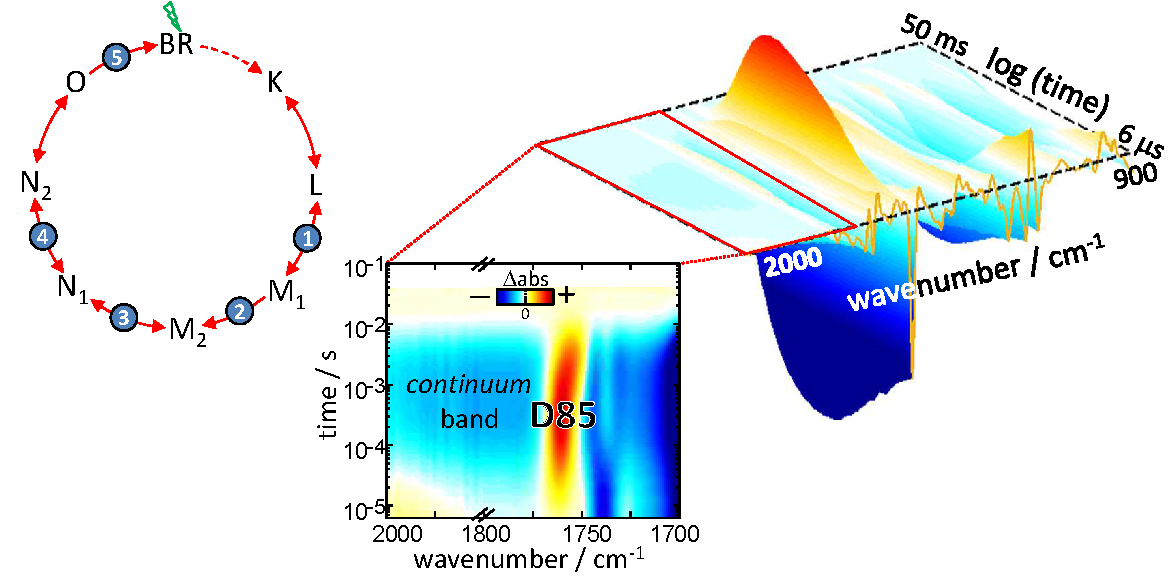
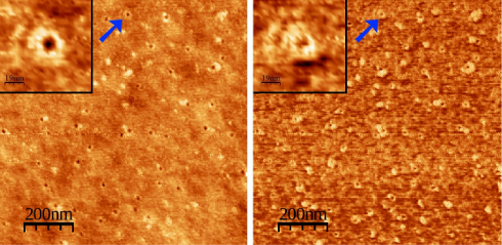
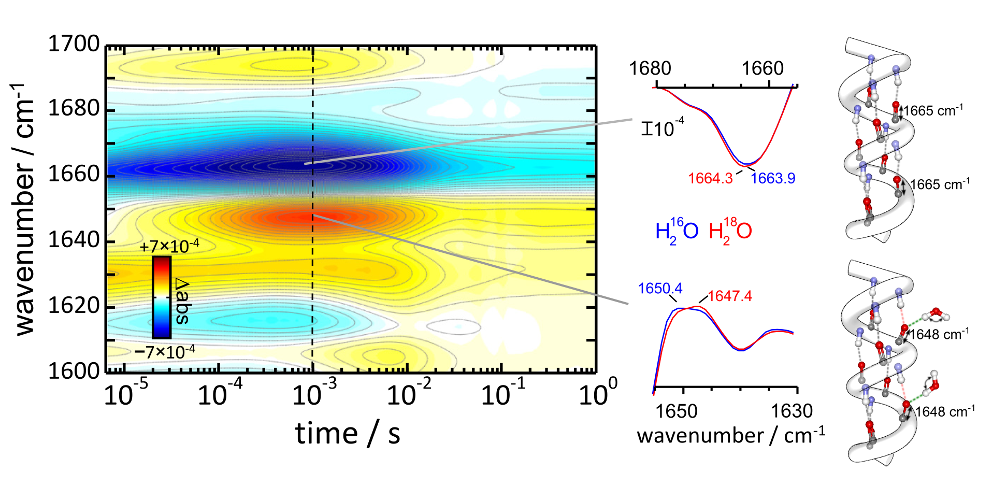
We investigate structural and dynamical aspects of membrane proteins and peptides, like folding, active transport and pore formation. We work as well in the optical control of proteins and peptides. Find out more in Projects.
Join us !
Postdocs, PhD students, Master's students and Undergraduate students are welcome to work in our group. Please see open calls and other opportunities to join MemBioPhys and contact us for any inquire.