-
Dom34 Links Translation to Protein O-mannosylation
van Wijlick L, Geissen R, Hilbig JS, Lagadec Q, Cantero PD, Pfeifer E, Juchimiuk M, Kluge S, Wickert S, Alepuz P, Ernst JF
(2016). ArticlePLoS Genet. No.2016 Oct 21;12(10):e1006395
Article Authors Metrics Comments Media Coverage Abstract Author Summary Introduction Results Discussion Methods and Materials Supporting Information Acknowledgments Author Contributions References Reader Comments Figures Abstract In eukaryotes, Dom34 upregulates translation by securing levels of activatable ribosomal subunits. We found that in the yeast Saccharomyces cerevisiae and the human fungal pathogen Candida albicans, Dom34 interacts genetically with Pmt1, a major isoform of protein O-mannosyltransferase. In C. albicans, lack of Dom34 exacerbated defective phenotypes of pmt1 mutants, while they were ameliorated by Dom34 overproduction that enhanced Pmt1 protein but...
Article Authors Metrics Comments Media Coverage Abstract Author Summary Introduction Results Discussion Methods and Materials Supporting Information Acknowledgments Author Contributions References Reader Comments Figures Abstract In eukaryotes, Dom34 upregulates translation by securing levels of activatable ribosomal subunits. We found that in the yeast Saccharomyces cerevisiae and the human fungal pathogen Candida albicans, Dom34 interacts genetically with Pmt1, a major isoform of protein O-mannosyltransferase. In C. albicans, lack of Dom34 exacerbated defective phenotypes of pmt1 mutants, while they were ameliorated by Dom34 overproduction that enhanced Pmt1 protein but not PMT1 transcript levels. Translational effects of Dom34 required the 5′-UTR of the PMT1 transcript, which bound recombinant Dom34 directly at a CA/AC-rich sequence and regulated in vitro translation. Polysomal profiling revealed that Dom34 stimulates general translation moderately, but that it is especially required for translation of transcripts encoding Pmt isoforms 1, 4 and 6. Because defective protein N- or O-glycosylation upregulates transcription of PMT genes, it appears that Dom34-mediated specific translational upregulation of the PMT transcripts optimizes cellular responses to glycostress. Its translational function as an RNA binding protein acting at the 5′-UTR of specific transcripts adds another facet to the known ribosome-releasing functions of Dom34 at the 3′-UTR of transcripts.
Read more Hide DOI: 10.1371/journal.pgen.1006395 -
Repositorio Datos: Gene expression omnibus (GEO)
Li T, De Clercq N, Medina DA, Garre E, Sunnerhagen P, Pérez-Ortín JE, Alepuz P
(2016). Recurs electrònicBiochim Biophys Acta.. No.1859(2):405-19
Acceso dataset: GSE72356. Tipo datos: Genomic Run On (RNA expresión.
-

The mRNA cap-binding protein Cbc1 is required for high and timely expression of genes by promoting the accumulation of gene-specific activators at promoters
Li T, De Clercq N, Medina DA, Garre E, Sunnerhagen P, Pérez-Ortín JE, Alepuz P.
(2016). ArticleBiochim Biophys Acta. No.2016 Feb;1859(2):405-19
The highly conserved Saccharomyces cerevisiae cap-binding protein Cbc1/Sto1 binds mRNA co-transcriptionally and acts as a key coordinator of mRNA fate. Recently, Cbc1 has also been implicated in transcription elongation and pre-initiation complex (PIC) formation. Previously, we described Cbc1 to be required for cell growth under osmotic stress and to mediate osmostress-induced translation reprogramming. Here, we observe delayed global transcription kinetics in cbc1Δ during osmotic stress that correlates with delayed recruitment of TBP and RNA polymerase II to osmo-induced promoters. Interestingly, we detect an interaction between Cbc1 and the MAPK Hog1, which controls most gene expression...
The highly conserved Saccharomyces cerevisiae cap-binding protein Cbc1/Sto1 binds mRNA co-transcriptionally and acts as a key coordinator of mRNA fate. Recently, Cbc1 has also been implicated in transcription elongation and pre-initiation complex (PIC) formation. Previously, we described Cbc1 to be required for cell growth under osmotic stress and to mediate osmostress-induced translation reprogramming. Here, we observe delayed global transcription kinetics in cbc1Δ during osmotic stress that correlates with delayed recruitment of TBP and RNA polymerase II to osmo-induced promoters. Interestingly, we detect an interaction between Cbc1 and the MAPK Hog1, which controls most gene expression changes during osmostress, and observe that deletion of CBC1 delays the accumulation of the activator complex Hot1–Hog1 at osmostress promoters. Additionally, CBC1 deletion specifically reduces transcription rates of highly transcribed genes under non-stress conditions, such as ribosomal protein (RP) genes, while having low impact on transcription of weakly expressed genes. For RP genes, we show that recruitment of the specific activator Rap1, and subsequently TBP, to promoters is Cbc1-dependent. Altogether, our results indicate that binding of Cbc1 to the capped mRNAs is necessary for the accumulation of specific activators as well as PIC components at the promoters of genes whose expression requires high and rapid transcription.
Read more Hide DOI: 10.1016/j.bbagrm.2016.01.002 -
Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae.
Li T, Belda-Palazón B, Ferrando A, Alepuz P*.
(2014). ArticleGenetics. No.Volume 197, Issue 4 (p.1191–120)
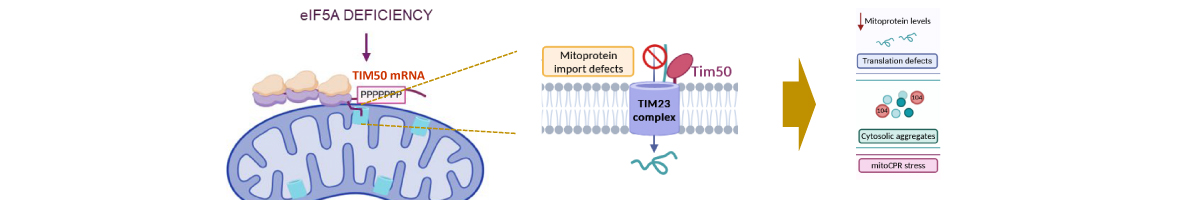
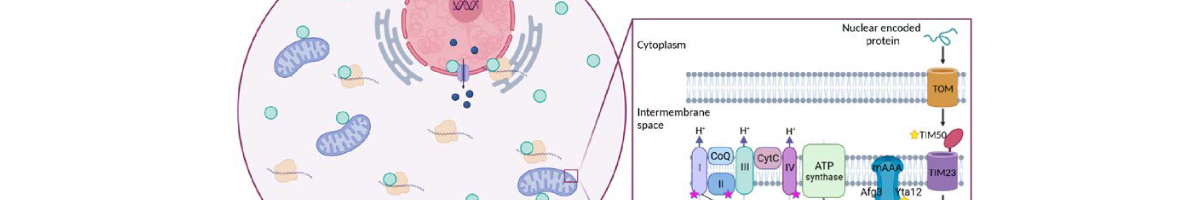
eIF5A is an essential and evolutionary conserved translation elongation factor, which has recently been proposed to be required for the translation of proteins with consecutive prolines. The binding of eIF5A to ribosomes occurs upon its activation by hypusination, a modification that requires spermidine, an essential factor for mammalian fertility that also promotes yeast mating. We show that in response to pheromone, hypusinated eIF5A is required for shmoo formation, localization of polarisome components, induction of cell fusion proteins, and actin assembly in yeast. We also show that eIF5A is required for the translation of Bni1, a proline-rich formin involved in polarized growth during...
eIF5A is an essential and evolutionary conserved translation elongation factor, which has recently been proposed to be required for the translation of proteins with consecutive prolines. The binding of eIF5A to ribosomes occurs upon its activation by hypusination, a modification that requires spermidine, an essential factor for mammalian fertility that also promotes yeast mating. We show that in response to pheromone, hypusinated eIF5A is required for shmoo formation, localization of polarisome components, induction of cell fusion proteins, and actin assembly in yeast. We also show that eIF5A is required for the translation of Bni1, a proline-rich formin involved in polarized growth during shmoo formation. Our data indicate that translation of the polyproline motifs in Bni1 is eIF5A dependent and this translation dependency is lost upon deletion of the polyprolines. Moreover, an exogenous increase in Bni1 protein levels partially restores the defect in shmoo formation seen in eIF5A mutants. Overall, our results identify eIF5A as a novel and essential regulator of yeast mating through formin translation. Since eIF5A and polyproline formins are conserved across species, our results also suggest that eIF5A-dependent translation of formins could regulate polarized growth in such processes as fertility and cancer in higher eukaryotes.
Read more HidePMID: 24923804
*corresponding author
DOI: 10.1534/genetics.114.166926 -

External conditions inversely change the RNA polymerase II elongation rate and density in yeast
Miguel A, Montón F, Li T, Gómez-Herreros F, Chávez S, Alepuz P, Pérez-Ortín JE.
(2013). ArticleBiochim Biophys Acta. No.2013 Nov;1829(11):1248-55
Elongation speed is a key parameter in RNA polymerase II (RNA pol II) activity. It affects the transcription rate, while it is conditioned by the physicochemical environment it works in at the same time. For instance, it is well-known that temperature affects the biochemical reactions rates. Therefore in free-living organisms that are able to grow at various environmental temperatures, such as the yeast Saccharomyces cerevisiae, evolution should have not only shaped the structural and functional properties of this key enzyme, but should have also provided mechanisms and pathways to adapt its activity to the optimal performance required. We studied the changes in RNA pol II elongation speed...
Elongation speed is a key parameter in RNA polymerase II (RNA pol II) activity. It affects the transcription rate, while it is conditioned by the physicochemical environment it works in at the same time. For instance, it is well-known that temperature affects the biochemical reactions rates. Therefore in free-living organisms that are able to grow at various environmental temperatures, such as the yeast Saccharomyces cerevisiae, evolution should have not only shaped the structural and functional properties of this key enzyme, but should have also provided mechanisms and pathways to adapt its activity to the optimal performance required. We studied the changes in RNA pol II elongation speed caused by alternations in growth temperature in yeast to find that they strictly follow the Arrhenius equation, and that they also provoke an almost inverse proportional change in RNA pol II density within the optimal growth temperature range (26-37 °C). Moreover, we discovered that yeast cells control the transcription initiation rate by changing the total amount of available RNA pol II
Read more HideErratum in:
Corrigendum to "External conditions inversely change the RNA polymerase II elongation rate and density in yeast" [Biochim. Biophys. Acta 1829/11 (2013) 1248-1255]. Miguel A, Montón F, Li T, Gómez-Herreros F, Chávez S, Alepuz P, Pérez-Ortín JE.Biochim Biophys Acta Gene Regul Mech. 2017 Feb;1860(2):289. doi: 10.1016/j.bbagrm.2016.11.003. Epub 2016 Dec 24.PMID: 27875711
DOI: 10.1016/j.bbagrm.2013.09.008 -

Dissection of the elements of osmotic stress response transcription factor Hot1 involved in the interaction with MAPK Hog1 and in the activation of transcription
Gomar-Alba M, Alepuz P, del Olmo Ml.
(2013). ArticleBiochim Biophys Acta. No.2013 Oct;1829(10):1111-25
The response to hyperosmotic stress is mediated by the HOG pathway. The MAP kinase Hog1 activates several transcription factors, regulates chromatin-modifying enzymes and, through its interaction with RNA polymerase II, it directs this enzyme to osmotic stress-controlled genes. For such targeting, this kinase requires the interaction with transcription factors Hot1 and Sko1. However, phosphorylation of these proteins by Hog1 is not required for their functionality. In this study, we aim to identify the Hot1 elements involved in Hog1-binding and in the activation of transcription. Two-hybrid experiments demonstrated that the Hot1 sequence between amino acids 340 and 534 and the CD element of...
The response to hyperosmotic stress is mediated by the HOG pathway. The MAP kinase Hog1 activates several transcription factors, regulates chromatin-modifying enzymes and, through its interaction with RNA polymerase II, it directs this enzyme to osmotic stress-controlled genes. For such targeting, this kinase requires the interaction with transcription factors Hot1 and Sko1. However, phosphorylation of these proteins by Hog1 is not required for their functionality. In this study, we aim to identify the Hot1 elements involved in Hog1-binding and in the activation of transcription. Two-hybrid experiments demonstrated that the Hot1 sequence between amino acids 340 and 534 and the CD element of Hog1 are required for the interaction between the two proteins and the Hot1-dependent transcription regulation. Inside this Hot1 region, short sequence KRRRR (KR4, amino acids 381–385) is essential for the kinase binding. Our data show that another element, sequence EDDDDD (ED5, amino acids 541–546), is essential for Hot1 binding to chromatin. Under osmotic stress conditions, both Hot1 elements, Hog1-interaction KR4 and DNA-binding ED5, are involved in the appropriate recruitment of Hog1 and RNA polymerase II to genes controlled by this transcription factor. Moreover, both sequences are required for osmotolerance and KR4 is necessary for the functionality of the HOG pathway. According to several experiments described in this study, the Hot1 protein is capable of forming homodimers.
Read more Hide DOI: 10.1016/j.bbagrm.2013.07.009 -

Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression [Review]
Pérez-Ortín JE, Alepuz P, Chávez S, Choder M.
(2013). ArticleJournal of Molecular Biology. No.2013 Oct 23;425(20):3750-75
mRNA concentration depends on the balance between transcription and degradation rates. On both sides of the equilibrium, synthesis and degradation show, however, interesting differences that have conditioned the evolution of gene regulatory mechanisms. Here, we discuss recent genome-wide methods for determining mRNA half-lives in eukaryotes. We also review pre- and posttranscriptional regulons that coordinate the fate of functionally related mRNAs by using protein- or RNA-based trans factors. Some of these factors can regulate both transcription and decay rates, thereby maintaining proper mRNA homeostasis during eukaryotic cell life.
DOI: 10.1016/j.jmb.2013.02.029 -
Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress
Garre E, Romero-Santacreu L, Barneo-Muñoz M, Miguel A, Pérez-Ortín JE, Alepuz P.
(2013). ArticlePLoS One. No.2013 Apr 19;8(4):e61240
The expression of ribosomal protein (RP) genes requires a substantial part of cellular transcription, processing and translation resources. Thus, the RP expression must be tightly regulated in response to conditions that compromise cell survival. In Saccharomyces cerevisiae cells, regulation of the RP gene expression at the transcriptional, mature mRNA stability and translational levels during the response to osmotic stress has been reported. Reprogramming global protein synthesis upon osmotic shock includes the movement of ribosomes from RP transcripts to stress-induced mRNAs. Using tiling arrays, we show that osmotic stress yields a drop in the levels of RP pre-mRNAs in S. cerevisiae...
The expression of ribosomal protein (RP) genes requires a substantial part of cellular transcription, processing and translation resources. Thus, the RP expression must be tightly regulated in response to conditions that compromise cell survival. In Saccharomyces cerevisiae cells, regulation of the RP gene expression at the transcriptional, mature mRNA stability and translational levels during the response to osmotic stress has been reported. Reprogramming global protein synthesis upon osmotic shock includes the movement of ribosomes from RP transcripts to stress-induced mRNAs. Using tiling arrays, we show that osmotic stress yields a drop in the levels of RP pre-mRNAs in S. cerevisiae cells. An analysis of the tiling array data, together with transcription rates data, shows a poor correlation, indicating that the drop in the RP pre-mRNA levels is not merely a result of the lowered RP transcription rates. A kinetic study using quantitative RT-PCR confirmed the decrease in the levels of several RP-unspliced transcripts during the first 15 minutes of osmotic stress, which seems independent of MAP kinase Hog1. Moreover, we found that the mutations in the components of the nonsense-mediated mRNA decay (NMD), Upf1, Upf2, Upf3 or in exonuclease Xrn1, eliminate the osmotic stress-induced drop in RP pre-mRNAs. Altogether, our results indicate that the degradation of yeast RP unspliced transcripts by NMD increases during osmotic stress, and suggest that this might be another mechanism to control RP synthesis during the stress response.
Read more Hide DOI: 10.1371/journal.pone.0061240 -
Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock
Garre E, Romero-Santacreu L, De Clercq N, Blasco-Angulo N, Sunnerhagen P, Alepuz P.
(2012). ArticleMolecular Biology of the Cell. No.Vol. 23, No. 1. pp.137-50.
In response to osmotic stress, global translation is inhibited, but the mRNAs encoding stress-protective proteins are selectively translated to allow cell survival. To date, the mechanisms and factors involved in the specific translation of osmostress-responsive genes in Saccharomyces cerevisiae are unknown. We find that the mRNA cap-binding protein Cbc1 is important for yeast survival under osmotic stress. Our results provide new evidence supporting a role of Cbc1 in translation initiation. Cbc1 associates with polysomes, while the deletion of the CBC1 gene causes hypersensitivity to the translation inhibitor cycloheximide and yields synthetic “sickness” in cells with limiting amounts of...
In response to osmotic stress, global translation is inhibited, but the mRNAs encoding stress-protective proteins are selectively translated to allow cell survival. To date, the mechanisms and factors involved in the specific translation of osmostress-responsive genes in Saccharomyces cerevisiae are unknown. We find that the mRNA cap-binding protein Cbc1 is important for yeast survival under osmotic stress. Our results provide new evidence supporting a role of Cbc1 in translation initiation. Cbc1 associates with polysomes, while the deletion of the CBC1 gene causes hypersensitivity to the translation inhibitor cycloheximide and yields synthetic “sickness” in cells with limiting amounts of translation initiator factor eIF4E. In cbc1Δ mutants, translation drops sharply under osmotic stress, the subsequent reinitiation of translation is retarded, and “processing bodies” containing untranslating mRNAs remain for long periods. Furthermore, osmostress-responsive mRNAs are transcriptionally induced after osmotic stress in cbc1Δ cells, but their rapid association with polysomes is delayed. However, in cells containing a thermosensitive eIF4E allele, their inability to grow at 37ºC is suppressed by hyperosmosis, and Cbc1 relocalizes from nucleus to cytoplasm. These data support a model in which eIF4E-translation could be stress-sensitive, while Cbc1-mediated translation is necessary for the rapid translation of osmostress-protective proteins under osmotic stress.
Read more Hide DOI: 10.1091/mbc.e11-05-0419 -

Global estimation of mRNA stability in yeast.
Marín-Navarro J, Jauhiainen A, Moreno J, Alepuz P, Pérez-Ortín JE, Sunnerhagen P.
(2011). ArticleMethods in Molecular Biology. No.2011;734:3-23
Turnover of mRNA is an important level of gene regulation. Individual mRNAs have different intrinsic stabilities. Moreover, mRNA stability changes dynamically with conditions such as hormonal stimulation or cellular stress. While accurate methods exist to measure the half-life of an individual transcript, global methods to estimate mRNA turnover have limitations in terms of resolution in time and precision. We describe and compare two complementary approaches to estimating global transcript stability: (1) direct measurement of decay rates; (2) indirect estimation of turnover from determination of mRNA synthesis rates and steady-state levels. Since the two approaches have distinct strengths...
Turnover of mRNA is an important level of gene regulation. Individual mRNAs have different intrinsic stabilities. Moreover, mRNA stability changes dynamically with conditions such as hormonal stimulation or cellular stress. While accurate methods exist to measure the half-life of an individual transcript, global methods to estimate mRNA turnover have limitations in terms of resolution in time and precision. We describe and compare two complementary approaches to estimating global transcript stability: (1) direct measurement of decay rates; (2) indirect estimation of turnover from determination of mRNA synthesis rates and steady-state levels. Since the two approaches have distinct strengths yet confer different cellular perturbations, it is valuable to consider results obtained with both methods. The practical aspects of the chapter are written from a yeast perspective; the general considerations hold true for all eukaryotes, however.
Read more Hide DOI: 10.1007/978-1-61779-086-7_1