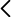CDEIGENT
CDEIGENT/2021/008
Title: Study of the importance of bacteriophages in the intestinal microbiome using single-cell genomics
Research group: Microbial Single-Cell Genomics
Clostridioides difficile infection (CDI) is an infection related to the use of broad-spectrum antibiotics that causes a perturbation of the composition of the intestinal microbiome, which allows C. difficile to initiate toxin production causing pseudomembranous colitis that can be fatal. CDI is an ideal model to study the relationships between bacteria and bacteriophages. There is a decrease in antibiotic treatment recommendations because of the ability of C. difficile to acquire antibiotic resistance linked to recurrent infections. This is why alternative experimental treatments are being explored, for example, specific bacteriophages or faecal microbiota transplantation. However, it remains unknown how the applied microorganisms (including bacteriophages) affect the composition of the patient’s gut microbiome.
The abundance of C. difficile in the intestinal microbiome is normally very low (<0.1%), suggesting that C. difficile bacteriophages are not necessarily detectable in metagenomic sequences or in the virus-enriched fraction (the virome) and are difficult to isolate using the traditional bacteriophage plaque methods. It is estimated that up to 15% of healthy individuals contain non-toxigenic C. difficile strains. If we apply single-cell genomics to the stool of CDI patients and healthy individuals carrying non-toxigenic C. difficile strains, we may detect bacteriophages on the surface of some C. difficile cells or replicating in their interior. It is a technique that has not yet been applied to any specific bacterial species (only the total proportion of specific bacteria attacked by bacteriophages has been investigated so far). It is not yet known whether CDI patients contain bacteriophages capable of attacking C. difficile or not. To answer this question, the proportion of C. difficile cells containing bacteriophages will be compared between healthy individuals and patients with CDI and, if some C. difficile bacteriophages are found in CDI patients, the genetic differences of bacteriophages and C. difficile strains in the two groups will be studied. The healthy individual containing the highest number of C. difficile bacteriophages will be used to perform an in vitro simulation of faecal microbiota transplantation in a test tube using the single-cell viral tagging method. We will investigate how bacteriophages from the healthy donor influence the bacterial composition of CDI patients.
single-cell genomics, metagenomics, C. difficile infection, bacteriophages
- Dzunkova -, Maria
- PI-Invest Disting Exper.Internacional
Generalitat Valenciana