Strengthening genomic surveillance of SARS-CoV-2 has been endorsed by the World Health Organization (WHO) to increase the capacity to detect new threats and achieve quality, timely and appropriate public health actions.
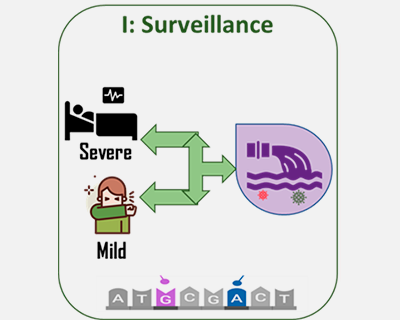
La detección de virus patógenos para humanos en aguas residuales es una herramienta muy útil y conocida para la vigilancia epidemiológica.
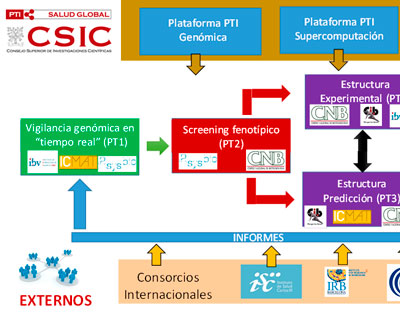
The objectives of this platform will be the identification of mutations associated with epidemiological/clinical changes of SARS-CoV-2 with special attention to breakthrough infections.
We are part of a joint effort to better understand the effects of mutations in SARS-CoV-2 in real time. This grant aims at building a platform to deal with current and future threats.
We are part of a joint effort to establish a platform for the screening of antivirals against current and future viral threats. The work combines groups that span the full drug discovery and development process.
SARS-CoV-2 is mainly a respiratory virus, although it’s been shown that it can replicate in the intestinal mucosa and be excreted via faecal matter. Molecular data proved the presence of the virus’s genetic material in faeces, while detection studies for infective viral particles are still incipient and based on a small number of patients.
The group of Dr. Mar Siles Lucas at IRNASA handles in vitro and in vivo models of parasitic-host interactions with Fasciola hepatica, which could be used to evaluate the potential of the parasite and its molecules to modulate relevant entry routes and inflammation routes in COVID-19. On one hand, this parasite has shown influence on the expression of endocytosis-related molecules (e.g. clathrins) on in vitro mouse epithelial cells, routes that may be relevant for SARS-Cov-2 entry to human cells. In addition, F. hepatica results in a modified Th2 response without the inflammatory component in its host in vivo, and this modulation may result in a controlled inflammatory response to COVID-19.
Defective interfering particles (DIP) are degenerate forms of viral genomes that are non-replicative but remain infectious by complementation with the wildtype (WT) virus. DIPs play a significant role in modulating the outcome of infection and immune responses, and can be artificially selected to strengthen their interfering activity and suppress replication of the full virus (therapeutic interfering particles, or TIPs). We propose to produce DIPs during SARS-CoV-2 replication in cell culture, purify them, test their antiviral effects, and characterize them molecularly. The outcome of the project will be a SARS-CoV-2 specific set of TIPs that could be used right away to treat COVID19.
Traditional viral infection diagnostic techniques in the clinic are based on (RT-)PCR procedures, which are time consuming and require precise equipment and human resources, which preclude a rapid and massive intervention. Here, we will engineer a novel class of stand-alone biosystems aimed at diagnosing the presence of SARS-CoV-2. These biosystems consist of three reaction steps following sample collection without the need of sophisticated equipment: i) isothermal amplification of viral RNA, ii) CRISPR-based nucleic acid detection (working like an on-the-fly sequencing reaction), and iii) output disclosure by immunochromatographic assay.
Although SARS-CoV-2 and other coronaviruses are primarily transmitted directly between individuals during epidemic phases, the permanence of the virus in the environment has the potential to cause new outbreaks despite mitigation efforts. SARS-CoV-1 was detected in hospital wastewater in China and it was shown that the viral particles could remain infective long enough to pose a risk.
This collaboration will employ a multi-disciplinary approach to identify inhibitors of SARS-CoV-2 receptor binding and entry. Combining expertise in medicinal chemistry, computational chemical biology, structural biology, and virology, 3 parallel approaches will be employed: (1) Immediate testing of compound libraries (>1,400) using a high-throughput cell assay for SARS-CoV-2 receptor binding and entry. (2) Computational screening and design, synthesis, and evaluation (3). Development of protein-based inhibitors derived from the ACE2 receptor. The final evaluation of hits for effectiveness and drug resistance will be done with SARS-CoV-2.
An ambitious scientific project led by researchers from the National Scientific Research Council (CSIC) in partnership with 40 hospitals all over Spain will study the compared genomes of the new coronavirus from patients with COVID-19 in order to understand and predict the evolution and epidemiology of the virus.
The project proposes a new strategy for the screening of drugs and antibodies against the SARS-CoV-2 coronavirus. This is an innovative technology for drug screening that uses an affordable, fast, safe and efficient system to evaluate all types of antiviral compounds and antibodies that block the entry of SARS-CoV-2 virus into human cells. The use of recombinant viruses has been successfully used in drug screening for various medically important viruses, as well as in serological screening. The advantages of the proposal are well founded, highlighting that this technology may be applicable in the future to other viruses that constitute an emerging public health problem.


 EvolCovid
EvolCovid
 PTI+: Transmission and contention
PTI+: Transmission and contention
 Genomic monitoring
Genomic monitoring
 Mutaciones
Mutaciones
 Antiviral platform
Antiviral platform
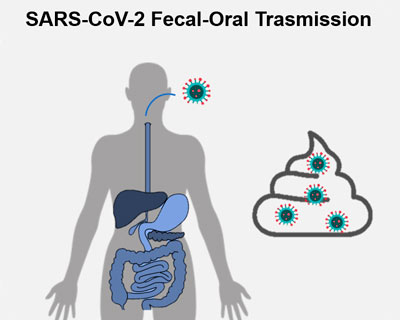 FecOrSARS
FecOrSARS
 COVID-19: anti-infectious and anti-inflammatory action of immunomodulatory parasite molecules in a safe-to-use synthetic format
COVID-19: anti-infectious and anti-inflammatory action of immunomodulatory parasite molecules in a safe-to-use synthetic format
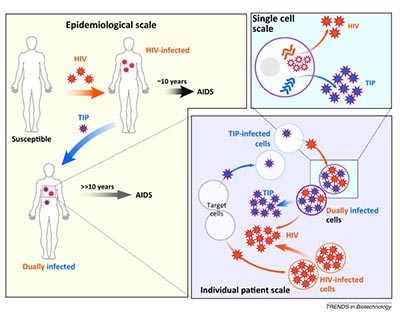 CoV2TIP
CoV2TIP
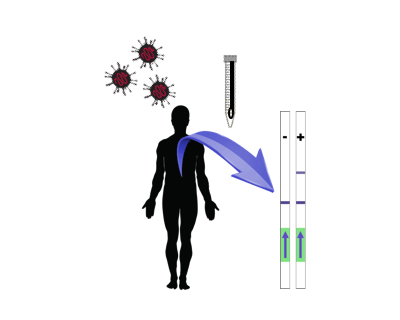 COV-CRISPIS
COV-CRISPIS
 EnvironmentalCOVID
EnvironmentalCOVID
 BlockAce
BlockAce
 SeqCOVID
SeqCOVID
 ANTICOR
ANTICOR