A team of researchers from Cavanilles Institute of Biodiversity and Evolutionary Biology, University of Valencia, led by Full-University Professor of Botany Eva Barreno, in collaboration with professors Eva del Campo and Leonardo Casano from the University of Alcalá, just published in the journal ‘Plos One’ the results of sequencing -by NGS techniques- chloroplast genome of strawberry plant (madroño in Spanish) which is a world innovation in regards to Mediterranean wildlife plants.
The strawberry tree (Arbutus unedo L.) is a shrub or small tree, with laurel and evergreen leaves living in Mediterranean habitats subject to certain episodes of mists. The genus Arbutus (Ericaceae, Ericales, Asterids, Dicotyledons, Angiosperms) comprises about 11 species, of which three are endemic to the Mediterranean, one to the Canary Islands and the rest are in California (USA). This area of separated distribution makes them considered as relict shrubs from the Tertiary and a possible origin of the genus in the vicinity of ancient sea Thetis is postulated, belonging to a floristic contingent biogeographically known as RAND-Flora, which could be adapted to Quaternary Mediterranean climate, where high temperatures coincide with low rainfall.
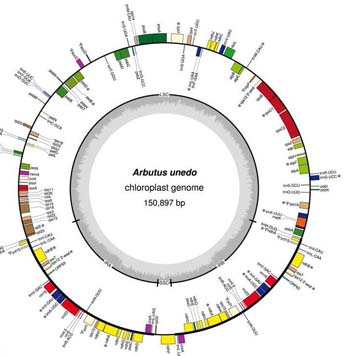
The group has found that the chloroplast genome (plastome) has gene losses, duplications and rearrangements, -due to significant investments- which, nevertheless, have maintained their average size -150.897 nucleotides- relative to other plants with flowers (angiosperms). Some of these changes may explain in part why some subtropical plants of the Tertiary flora could suit drastic drought conditions that marked the onset of the Mediterranean climate.
The plastome of vascular plants was originated from the acquisition, by endosymbiosis processes, of genome of a photosynthetic cyanobacterium within a eukaryotic cell, so their DNA is circular architecture is divided into four regions: a long copy single (LSC) and a single small copy (SSC) separated by a double zone called inverted-repeated (IR) of identical sequences located as a mirror image. The chloroplast arbutus shows, compared to the other sequenced angiosperms, a SSC region five times lower, or full loss of function (pseudogenization) of genes ycf1, ycf2, clpP, accD and rps16 essential for plant life. In addition, the gene complex ndhH-D has been relocated from the SSC region to the two IR -novelty for angiosperms- so there is a duplicate and is known to have essential functions to cope with oxidative stress.
It is also new the fact that long "tandem repetitions" are located in areas of rearrangements of genes and pseudogenes. Previously, it was thought that the plastid genomes of angiosperms were highly conserved in terms of the organization and composition of their genes, but this study emphasizes that the Ericaceae family represents a distinct evolutionary lineage from the rest of the other families of the order Ericales, underlining the paraphyletic nature of Ericales all the rest of angiosperms with fused petals (Asterids).
At the same time, within this work has been used plastome sequencing for phylogenetic analysis complementary to the rest of Asterids to solve some important uncertainties about the evolutionary relationships within this diverse group of flowering plants.
This research provides evidence of the originality of the plastid genome of the Ericales. Comparative analysis will be a valuable source of information on genomic restructuring events that occurred during the evolution of the Ericales. Furthermore, from an ecophysiological perspective, there is a consensus that chlorine breathing and ndh complex are essential to prevent oxidative stress from free radicals generated in the metabolism so that plants with this plastid structure will be better able to cope with uncertainties of current climate change and are undoubted future biotechnological applications in crops.
The research was conducted using the sequencing technology 454 in the company "Lifesequencing" of the Science Park of the University of Valencia. The arduous process of isolation of chloroplasts was performed with protocols designed by Julian Perez in 'Secugen'. For the study a village by the river Viejas (Montes de Toledo) was selected, where optimal habitats of its range are on the Iberian Peninsula.
Eva Barreno team, which has also sequenced the genome of symbiotic algae of lichens, belongs to the group 'Symbiosis, Diversity and Evolution in Plants and Lichens: Biotechnology and Innovation' of Cavanilles Institute of Biodiversity and Evolutionary Biology (ICBIBE) of the University of Valencia, to the Department for 'Life Sciences' at the University of Alcalá and the 'Biodiversity and Conservation' are of the Rey Juan Carlos University.
Reference article:
*MARTÍNEZ-ALBEROLA Fernando, DEL CAMPO Eva. M., LÁZARO-GIMENO David, MEZQUITA-CLARAMONTE Sergio, MOLINS, Arantxa, MATEU Isabel, PEDROLA-MONFORT Joan, CASANO Leonardo M.; BARRENO, Eva. (2013). Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts.
PLoS ONE 8(11): e79685. doi:10.1371/journal.pone.0079685
Last update: 1 de january de 2014 07:00.
News release